Laboratory of Programmable Polymers
Did you know that polymers can mimic functions displayed by natural macromolecules?
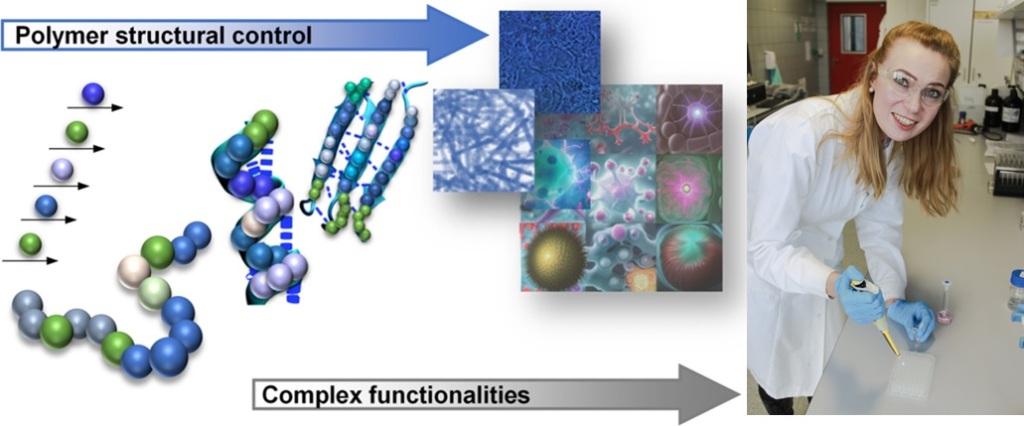
In nature, DNA and proteins with well-ordered structural skeletons can be recognized as sequence-defined macromolecules. DNA, composed of A, T, C, G nucleotides, is a natural polymer of two chains that coil around each other to form a double helix carrying genetic instructions. All essential informations to generate sequential protein chains from amino acids, by transcription and translation process, are encoded in the DNA sequence. Subsequently, the amino acid sequence determines three-dimensional structure of the protein and decides about their functions.
The synthesis of uniform macromolecules with defined monomer sequence, as displayed by natural polymers, is a challenge in modern polymer chemistry. In order to achieve full control over polymer structure, new synthesis strategies, based on iterative chemistry, has been recently developed. The accessibility of sequence-defined, uniform macromolecular structures enabled the design of polymeric materials beyond classical polymers applications, e.g. data storage. Furthermore, the monomer sequence regulation became an important parameter for the fine modulation of properties and functions of synthetic materials.
about Programmable Polymers
Our goal
Our ambition is to develop life-like functions in synthetic polymers through regulation of monomer sequence for development of complex materials.
Work With Us
We are always open for new, motivated people to join our team.
We are looking forward to collaborations with industrial partners to develop new technologies based on functional polymers.
Our work
Stay tuned about our research projects ranging from fundamental science to applied research and development. Follow us on social media and on our site to get updated on our progress.



